Before applying K-mean clustering Algorithm on Random data let me clear out some of the general terminologies that I will be using in this and upcoming tutorials, so let’s me briefly explain what they are:
What is Algorithm?
A set of instruction that must be followed when solving a particular problem is known as Algorithm
What is Clustering?
Clustering is a method to divide anything into a few groups, where elements of each group have more similarity to each other than those elements in other other groups. For example dividing mixture of fruit based on that type they are belong to i.e. Mango, Apple, Banana, etc.
What is Clustering Algorithm?
Clustering algorithm is an unsupervised machine learning task, involving automatically discovering natural grouping in data. Unlike supervised learning (like predictive modeling), clustering algorithms only interpret the input data and find natural groups or clusters in feature space.
There are so many Clustering Algorithm out there such as:
- K-Mean
- DBSCAN
- Mini-Batch -Means Mean Shift
- Gaussian Mixture Model
- OPTICS
- .etc
What is the K-Means Clustering Algorithm?
K-Means clustering algorithm group data into different different cluster based on the information we provided to the algorithm to perform clustering on the dataset. and it is one of the simplest unsupervised learning algorithms that solve the well known clustering problem i.e. in Health care sector it used to identify which medicine belong to which category and so more, I will be talking more about it in upcoming tutorials. The Algorithm follows a simple and easy way to classify a given data set through a certain number of clusters (assume k clusters). The main idea is to define k centers, one for each cluster.
which are usually chosen to be far enough apart from each other spatially, in Euclidean Distance, to be able to produce effective results. Each cluster has a center, known as centroid, and a data point is clustered into a certain cluster based on how close the features are to the centroid.
How the K-means algorithm work?
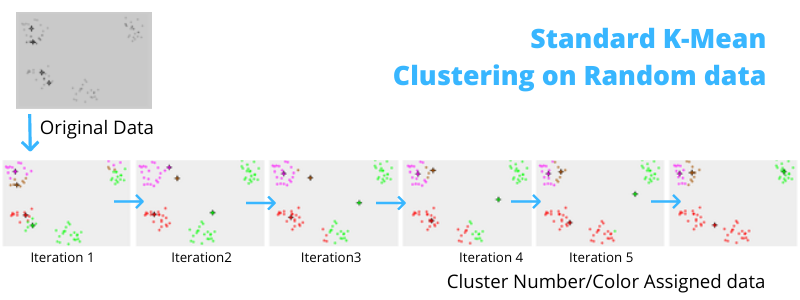
K-means algorithm iteratively minimizes the distances between every data point and its centroid in order to find the most optimal solution for all the data points.
- K random points are chosen to be centroids.
- Distances between every data point and the k centroids are calculated.
- Based on calculated distance, each point is assigned to the nearest cluster
- New cluster centroid positions are updated: similar to finding a mean in the point locations
- If the centroid locations changed, the process repeats from step 2, until the calculated new center stays the same, which signals that the clusters' members and centroids are now set. Here is a C implementation of k-mean for text file
Finding the minimal distances between all the points implies that data points have been separated in order to form the most compact clusters possible. In other words, no other iteration could have a lower average distance between the centroids and the data points found within them.
When to Use K-Means Clustering?
K-Means clustering is a fast, robust, and simple algorithm that gives reliable results when data sets are distinct or well separated from each other in a linear fashion. It is best used when the number of cluster centers is specified due to a well-defined list of types shown in the data. However, it is important to keep in mind that K-Means clustering may not perform well if it contains heavily overlapping data, or if the data is noisy or full of outliers.
Example: For Random Data
Now Let's create a new project by running commands-
cargo new kmean_using_linfa
This will create the project folder kmean_using_linfa, now navigate inside it by performing command:
cd kmean_using_linfa
Now add required dependency by updating Cargo.toml file as such:
Cargo.toml:
[package]
name = "kmean_using_linfa"
version = "0.1.0"
edition = "2021"
# See more keys and their definitions at https://doc.rust-lang.org/cargo/reference/manifest.html
[dependencies]
linfa = "0.5.0"
linfa-clustering = "0.5.0"
ndarray = "0.15.4"
ndarray-rand = "0.14.0"
rand_isaac = "0.3.0"
approx = "0.5.0"
and main.rs file as such:
use linfa::DatasetBase;
use linfa::traits::{Fit, Predict};
use linfa_clustering::{ KMeans, generate_blobs};
use ndarray::{Axis, array};
use ndarray_rand::rand::SeedableRng;
use rand_isaac::Isaac64Rng;
fn main() {
// Our random number generator, seeded for reproducibility
let seed = 42;
let mut rng = Isaac64Rng::seed_from_u64(seed);
// `expected_centroids` has shape `(n_centroids, n_features)` which will automatically get changed as stated in algorithm
// i.e. three points in the 2-dimensional plane
let expected_centroids = array![[0., 1.], [-10., 20.], [-1., 10.]];
println!("Initialized Centroid: {:?}\n",expected_centroids);
// Let's generate a synthetic dataset: three blobs of observations
// (100 points each) centered around our `expected_centroids`
let data = generate_blobs(100, &expected_centroids, &mut rng);
println!("Randomly Generated Data: {:?}\n",data);
let n_clusters = expected_centroids.len_of(Axis(0));
println!("Expected Number of Cluster: {:?}\n",n_clusters);
let observations = DatasetBase::from(data.clone());
// Let's configure and run our K-means algorithm
// We use the builder pattern to specify the hyperparameters
// `n_clusters` is the only mandatory parameter.
// If you don't specify the others (e.g. `n_runs`, `tolerance`, `max_n_iterations`)
// default values will be used.
let model = KMeans::params_with_rng(n_clusters, rng.clone())
.tolerance(1e-2)
.fit(&observations)
.expect("KMeans fitted");
println!("Resultant Centroid: {:?}",model.centroids());
// Once we found our set of centroids, we can also assign new points to the nearest cluster
let new_observation = DatasetBase::from(array![[20., 20.5]]);
// Predict returns the **index** of the nearest cluster
let dataset = model.predict(new_observation);
// We can retrieve the actual centroid of the closest cluster using `.centroids()`
let closest_centroid = &model.centroids().index_axis(Axis(0), dataset.targets()[0]);
println!("Closest Centroid of new input data {} : {}\n",array![20., 20.5], closest_centroid);
}
To build the project run command:
cargo build
and to run the project run command:
cargo run
And we are done! The output will be something like this:
ajayrmavs33236@rmavs:~/Desktop/kmean_using_linfa$ cargo run
Finished dev [unoptimized + debuginfo] target(s) in 0.04s
Running `target/debug/kmean_using_linfa`
Initialized Centroid: [[0.0, 1.0],
[-10.0, 20.0],
[-1.0, 10.0]], shape=[3, 2], strides=[2, 1], layout=Cc (0x5), const ndim=2
Randomly Generated Data: [[-0.11236555285602912, 1.7007428833554714],
[0.8950533285467516, 0.6025078239189654],
[-0.13018258473931488, 1.8638228493554734],
[1.2657209367270663, 1.267501999871629],
[0.5761781023204727, 2.0698819932798553],
...,
[-1.4744152739536533, 9.795872353678346],
[-1.5496360174973098, 10.0789731813974],
[-1.3066330855716237, 11.420941486545386],
[-1.5502955676043042, 9.503241021092913],
[-2.227387210297529, 11.276716979963245]], shape=[300, 2], strides=[2, 1], layout=Cc (0x5), const ndim=2
Expected Number of Cluster: 3
Resultant Centroid: [[-10.072365022783869, 19.976063571297026],
[-1.0678160111577655, 9.911730944101146],
[0.01580578275521944, 1.0495069733592552]], shape=[3, 2], strides=[2, 1], layout=Cc (0x5), const ndim=2
Closest Centroid of new input data [20, 20.5] : [-1.0678160111577655, 9.911730944101146]
Also to produce a Webassembly file run command
cargo wasi build --release or cargo build --release --target=wasm32-wasi
and to run this project inside TEE instances such as Enarx Keeps
follow these instruction, As this is related to data, so next I will be talking about how Confidential Computing works and how we can achieve the same with Enarx and see examples for the same.
Link to previous tutorial can be found over here



Top comments (0)